last update: 02/27/2007

| Input format |
ESEfinder accept sequences in the FASTA format. A sequence in FASTA format begins with a ">" and then a single line description, followed by lines sequence data. Multiple sequences are allowed and each one should have a descriptive line. An example of a FASTA file is given below:
>seq1 aaatagacaattttaaaaaccaaacagaatgggactgtcttctgcaagcc tacctacaaacag > aaatcagttcgtcttcgttgcagcagatgaaatttcttgtaacacctctg cag
The query sequences can be directly pasted into the text box, or can be uploaded from a text file
| Size limit |
| Output format |
ESEfinder 3.0 has made a number changes in the output format.
In the first page, the user can choose to generate output in either html format or plain text format.
In html format, the result of each sequence is displayed separately, each has three sections
- sequence summary
- tabular output
- graphical output
Since release 2.0, ESEfinder allows to direct graphical display of the results
- the analyzed (exonic) sequence is reproduced along
the X-axis.
Either the actual nucleotides or the nucleotide number can be displayed - only the high-score values (the values above the selected threshold) are represented
- the height of the bars represents the motif scores
- the width of the bars indicates the length of the motif (6, 7 or 8 nucleotides)
- the different SR protein high-scores are color-coded
as illustrated below
| An example |
>BRCA1 WT
ATGCTGAGTTTGTGTGTGAACGGACACTGAAATATTTTCTAGGAATTGCGGGAGGAAAATGGGTAGTTAGCTATTTCT
| seq #1 | id: BRCA1 WT |
length:78 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Sequence ATGCTGAGTTTGTGTGTGAACGGACACTGAAATATTTTCTAGGAATTGCGGGAGGAAAATGGGTAGTTAGCTATTTCT Tabular results
Graphical results 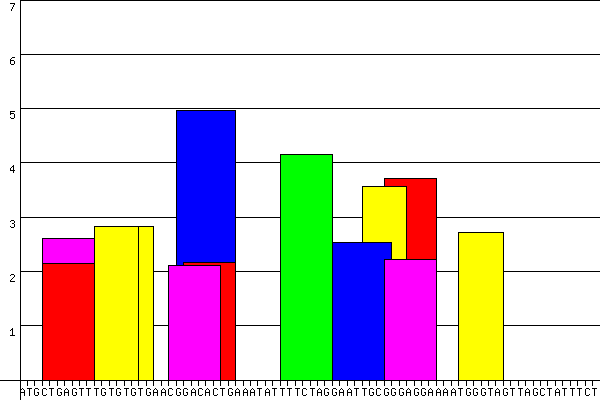
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Krainer Lab and
Zhang Lab,
Cold Spring Harbor Laboratory, all
rights reserved
Questions/suggestions email: Jinhua Wang.