last update: 02/27/2007

A typical mammalian gene is composed of several relatively short exons that are interrupted by much longer introns. To generate correct mature mRNAs, the exons must be identified and joined together precisely and efficiently, in a process that requires the coordinated action of five small nuclear (sn)RNAs (U1, U2, U4, U5 and U6) and more than 60 polypeptides. The inaccurate recognition of exon/intron boundaries or the failure to remove an intron generates aberrant mRNAs that are either unstable or code for defective or deleterious protein isoforms.
Paradoxically, in higher eukaryotes, the requirement for accurate splicing is met by exon-intron junctions that are defined by weakly conserved intronic cis-elements: the 5' splice site, 3' splice site and branch site. These elements are necessary but by no means sufficient to define exon/intron boundaries.
Several examples of intronic and exonic cis-elements that are important for correct splice-site identification and are distinct from the classical splicing signals have been described. These elements can act both by stimulating (enhancers) or by repressing (silencers) splicing, and they seem to be especially relevant for regulating alternative splicing. Exonic splicing enhancers (ESEs), in particular, appear to be very prevalent, and may be present in most, if not all exons, including constitutive ones
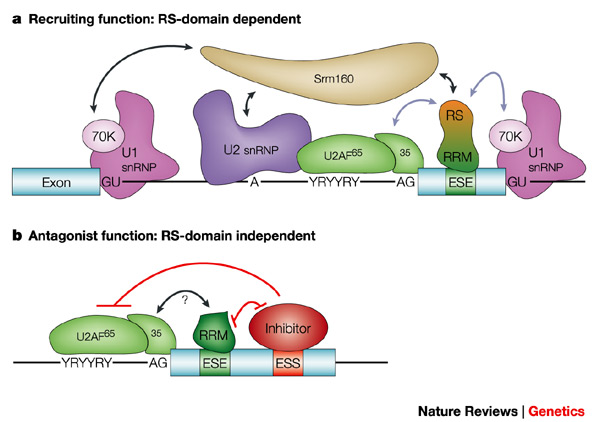
Exonic enhancers are thought to serve as binding sites for specific serine/arginine-rich (SR) proteins, a family of structurally related and highly conserved splicing factors characterized by one or two RNA-recognition motifs (RRM) and by a distinctive C-terminal domain highly enriched in RS dipeptides (the RS domain). The RRMs mediate sequence-specific binding to the RNA, and so determine substrate specificity, whereas the RS domain appears to be involved mainly in protein-protein interactions. SR proteins bound to ESEs can promote exon definition by directly recruiting the splicing machinery through their RS domain and/or by antagonizing the action of nearby silencer elements.
|
|
|
Models of SR protein
action in exonic-splicing-enhancer-dependent splicing
a)RS-domain-dependent mechanism. An SR protein
binds to an ESE through its RRMs and contacts the splicing factor U2AF35
and/or the snRNP protein U1-70K at the adjacent splice sites through its
RS domain. The large subunit of U2AF (U2AF65) binds to the
polypyrimidine (Y) tract, which here is interrupted by purines (R) and
is therefore part of a weak 3' splice site. U2AF65 also promotes binding
of U2 snRNP to the branch site. U2AF35 recognizes the 3' splice-site AG
dinucleotide. The U1 snRNP particle binds to the upstream and downstream
5' splice sites through base paring of the U1 snRNA; the 70K polypeptide
of each U1 snRNP particle is shown. The three sets of splicing-factor:pre-mRNA
interactions (U2AF:3' splice site, U1 snRNP:5' splice site and SR
protein:ESE) are strengthened by the protein:protein interactions (blue
arrows) that are mediated by the RS domain. For some ESE-dependent
pre-mRNAs, indirect interactions (black arrows) are bridged by the
splicing co-activator Srm160, which stimulates splicing of some
ESE-dependent pre-mRNAs and also interacts with the U2 snRNP. |
Selected references:
Cartegni L., Chew S.L., & Krainer A.R.
Listening to silence and understanding nonsense: exonic mutations that
affect splicing. Nat Rev Genet. 2002 3(4),285-98.
Caceres JF, Kornblihtt AR.
Alternative splicing: multiple control mechanisms and involvement in
human disease.Trends Genet. 18(4), 186-93. (2002).
Hastings, M.L. & Krainer, A.R.
Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol
13, 302-9. (2001).
Graveley, B.R.
Alternative splicing: increasing diversity in the proteomic world.
Trends Genet 17, 100-7. (2001).
Blencowe, B.J.
Exonic splicing enhancers: mechanism of action, diversity and role in
human genetic diseases. Trends Biochem Sci 25, 106-10.
(2000).
Graveley, B.R.
Sorting out the complexity of SR protein functions. Rna 6,
1197-211. (2000).
Krainer Lab and
Zhang Lab,
Cold Spring Harbor Laboratory, all
rights reserved
Questions/suggestions email: Jinhua Wang.